Institute of Biomedical Research and InnovationDepartment of Hematology-Oncology
Research Overview
Understanding and Targeting the Dysregulated Pathways in Malignant or Aged Hematopoiesis
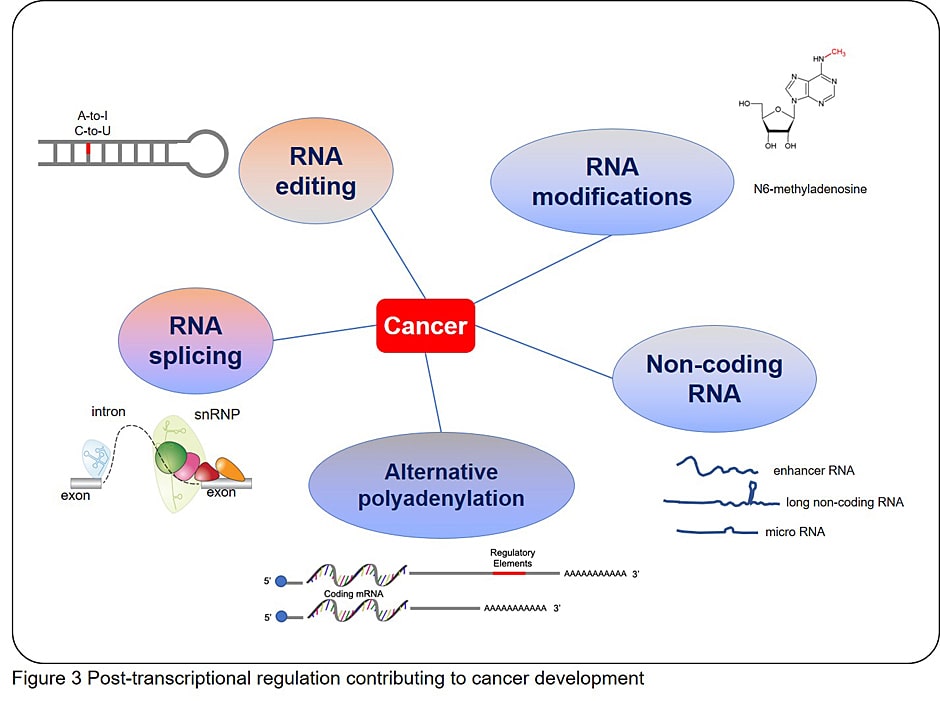
We pursue undetermined mechanisms for developing and sustaining malignant and/or aged hematopoiesis, mainly focusing on post-transcriptional regulation (e.g., pre-mRNA splicing, RNA modification, RNA transport, and translation initiation), cancer-specific metabolic pathways (e.g., ferroptosis), and extrinsic support by the bone marrow (BM) microenvironment. Using novel animal models, high-throughput screening, and patient-derived data, we will explore here-to-fore unrecognized characteristics of malignant and/or aged hematopoiesis and develop a mechanism-based therapeutic approach for malignant and/or aged hematopoiesis.
NEWS
Member
- Professor
-
Research Scientist
-
Masaki Nomura
-
-
Research Assistant
-
Miki Fukumoto
-
Yui Koike
-
Hiromi Ito
-
-
Graduate Student
-
Atsushi Tanaka
-
Muran Xiao
-
Yifan Zhang
-
Weijia Zang
-
Yumi Aoyama
-
Wataru Saika
-

- Research focus
- Publications
Research focus
Cancer cells depend on differential programs for their proliferation, metabolism, and self-renewal than normal cells. Although these are partially attributed to genetic mutations in cancer cells, precise mechanisms largely remain unsolved. In addition, intrinsic and extrinsic pathways are necessary to sustain cancer stemness. Here, we aim to better understand cancers (mainly myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML)), particularly focusing on the following aspects:
- (1) Oncogenic mechanisms based on aberrant post-transcriptional regulation
- - Determine the role of BRD9 in normal and malignant hematopoiesis
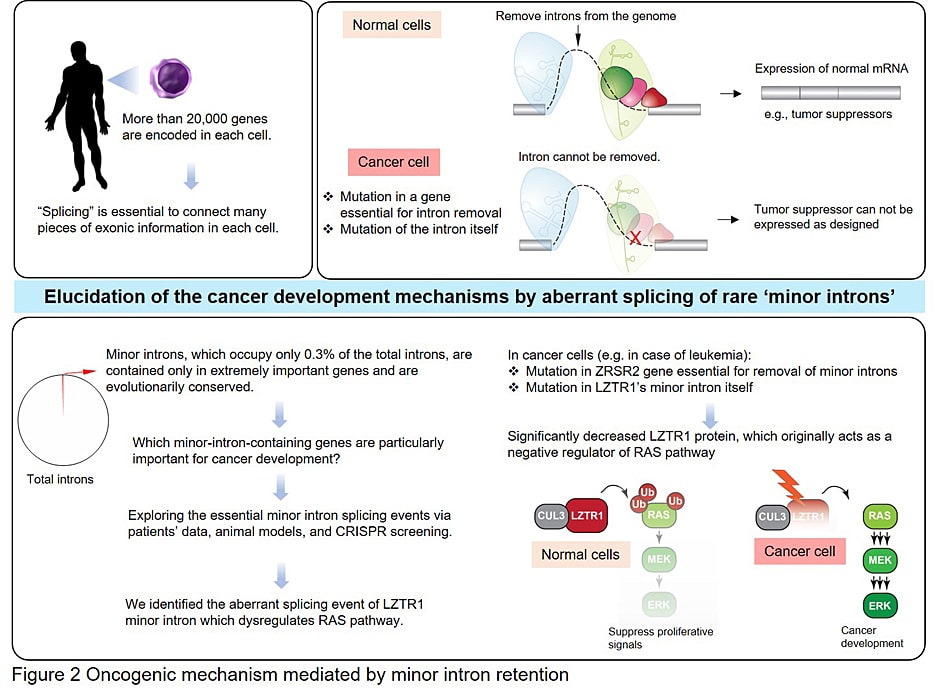
- - Elucidate the functional connection between minor intron splicing and stemness
- - Clarify how malignant hematopoietic stem cells (HSCs) depend on distinct RNA methylation and transport system
- - Explore the therapeutic strategies against RNA methylation enzymes and Nuclear Pore Complex (NPC) in AML
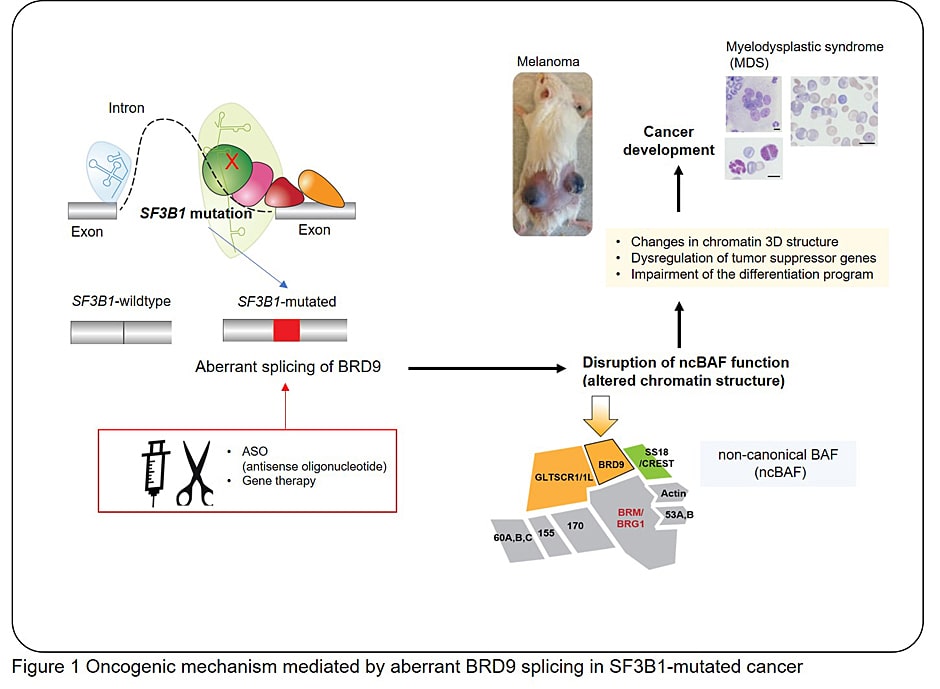
- (2) Targeting BRD9 for AML treatment and developing a synthetic lethal approach in SF3B1-mutated cancer
- (3) Generation of pre-clinical models of poor-prognostic leukemia to elucidate the underlying mechanisms
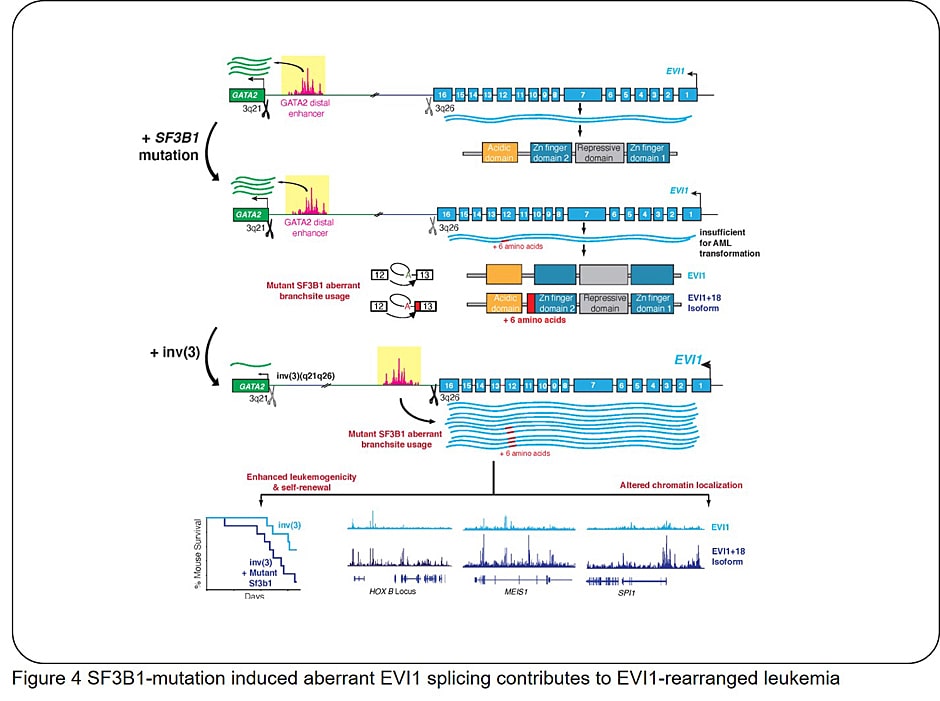
- (4) Clarify how impaired HSCs and BM microenvironment affect each other
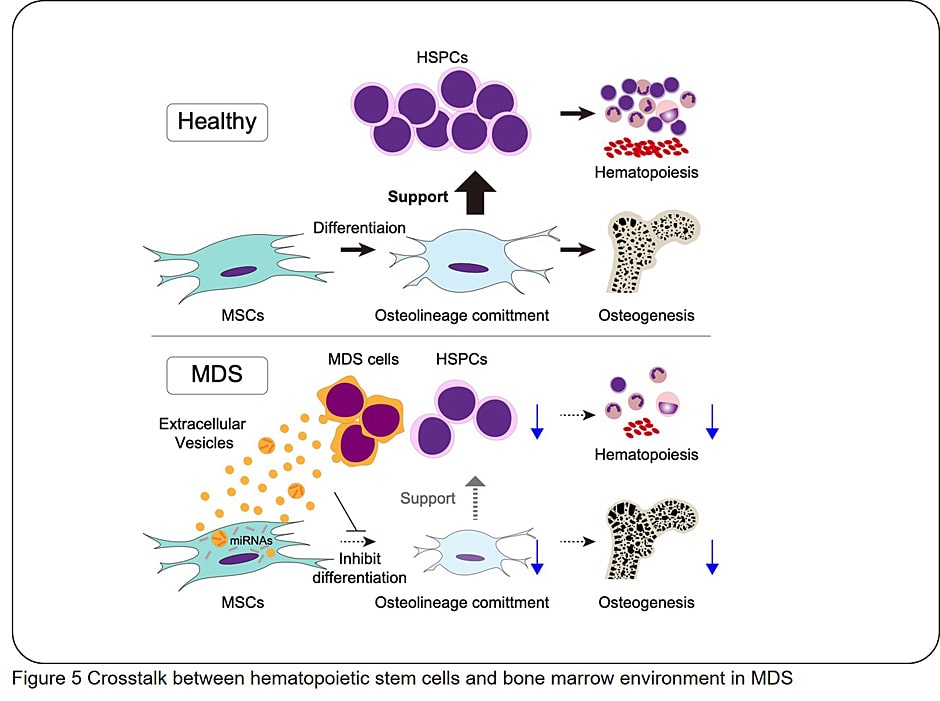
- (5) Understanding biological effects of "ferroptosis" in AML cells and aged BM for novel therapies
Publications and Activities
- Chen S, Vedula RS, Cuevas-Navarro A, Lu B, Hogg SJ, Wang E, Benbarche S, Knorr K, Kim WJ, Stanley RF, Cho H, Erickson C, Singer M, Cui D, Tittley S, Durham BH, Pavletich TS, Fiala E, Walsh MF, Inoue D, Monette S, Taylor J, Rosen N, McCormick F, Lindsley RC, Castel P, Abdel-Wahab O. Impaired proteolysis of non-canonical RAS proteins drives clonal hematopoietic transformation. Cancer Discov. 2022 Oct 5;12(10):2434-2453.
- Nishimura K, Yamazaki H, Zang W, Inoue D. Dysregulated Minor Intron Splicing in Cancer. Cancer Sci. 2022 Sep;113(9):2934-2942.
- Tanaka A, Nakano AT, Nomura M, Yamazaki H, Bewersdorf JP, Lazaro RM, Hogg S, Liu B, Penson A, Yokoyama A, Zang W, Havermans M, Koizumi M, Hayashi Y, Cho H, Kanai A, Lee SC, Xiao M, Koike Y, Zhang Y, Fukumoto M, Aoyama Y, Konuma T, Kunimoto H, Inaba T, Nakajima H, Honda H, Kawamoto H, Delwel R, Abdel-Wahab O, Inoue D. Aberrant EVI1 splicing contributes to EVI1-rearranged leukemia. Blood, 2022 Jun 16:blood.2021015325. doi: 10.1182/blood.2021015325. Online ahead of print.
- Tanaka A, Nakano AT, Nomura M, Yamazaki H, Bewersdorf JP, Lazaro RM, Hogg S, Liu B, Penson A, Yokoyama A, Zang W, Havermans M, Koizumi M, Hayashi Y, Cho H, Kanai A, Lee SC, Xiao M, Koike Y, Zhang Y, Fukumoto M, Aoyama Y, Konuma T, Kunimoto H, Inaba T, Nakajima H, Honda H, Kawamoto H, Delwel R, Abdel-Wahab O, Inoue D. Aberrant EVI1 splicing contributes to EVI1-rearranged leukemia. Blood, 2022 Aug 25;140(8):875-888.
- Inoue D, Polaski JT, Taylor J, Castel P, Chen S, Kobayashi S, Hogg SJ, Hayashi Y, Bello Pineda JM, Ettaib EM, Erickson C, Knorr K, Fukumoto M, Yamazaki H, Tanaka A, Fukui C, Lu XL, Durham BH, Liu B, Wang E, Mehta S, Zakheim D, Grippa R, Penson A, Chew GL, McCormick F, Bradley RK, Abdel-Wahab O. Minor intron retention drives clonal hematopoietic disorders and diverse cancer predisposition. Nat Genetics, 2021 May;53(5):707-718.
- Rahmani NE, Ramachandra N, Sahu S, Gitego N, Lopez A, Pradhan K, Bhagat TD, Gordon-Mitchell S, Pena BR, Kazemi M, Rao K, Giricz O, Maqbool SB, Olea R, Zhao Y, Zhang J, Dolatshad H, Tittrea V, Tatwavedi D, Singh S, Lee J, Sun T, Steidl U, Shastri A, Inoue D, Abdel-Wahab O, Pellagatti A, Gavathiotis E, Boultwood J, Verma A. Blood Cancer J, 2021 Sep 21;11(9):157.
- Trivedi G, Inoue D, Zhang L. Targeting low-risk myelodysplastic syndrome with novel therapeutic strategies. Trends Mol Med. 2021 Oct;27(10):990-999.
- Fujino T, Goyama S, Sugiura Y, Inoue D, Asada S, Yamasaki S, Matsumoto A, Yamaguchi K, Isobe Y, Tsuchiya A, Shikata S, Sato N, Morinaga H, Fukuyama T, Tanaka Y, Fukushima T, Takeda R, Yamamoto K, Honda H, Nishimura E, Furukawa Y, Shibata T, Abdel-Wahab O, Suematsu M, Kitamura T. Mutant ASXL1 induces age-related expansion of phenotypic hematopoietic stem cells through activation of Akt/mTOR pathway. Nat Commun, Mar 23;12(1):1826. doi: 10.1038/s41467-021-22053-y..
- HHEX promotes myeloid transformation in cooperation with mutant ASXL1. Takeda R, Asada S, Park SJ, Yokoyama A, Becker HJ, Kanai A, Visconte V, Hershberger CE, Hayashi Y, Yonezawa T, Tamura M, Fukushima T, Tanaka Y, Fukuyama T, Matsumoto A, Yamasaki S, Nakai K, Yamazaki S, Inaba T, Shibata T, Inoue D, Honda H, Goyama S, Maciejewski JP, Kitamura T. Blood, 2020 Oct 1;136(14):1670-1684.
- Inoue D, Chew GL, Liu B, Michel BC, Pangallo J, D’Avino AR, Hitchman T, North K, Lee SC, Bitner L, Ariele B, Moore AR, Yoshimi A, Hoyos LE, Cho H, Penson A, Lu SX, Taylor J, Chen Y, Kadoch C, Abdel-Wahab O, Bradley RK. Spliceosomal disruption of the non-canonical BAF complex in cancer. Nature, 2019 Oct;574(7778):432-436.
- Yoshimi A, Lin KT, Wiseman DH, Rahman MA, Pastore1 A, Intlekofer AM, Wang B, Lee SCW, Micol JB, Zhang XJ, Inoue D, Thompson CB, Levine RL, Bradley RK, Abdel-Wahab O. Coordinated Alterations in RNA Splicing and Epigenetic Regulation Drive Leukemogenesis. Nature, 2019 Oct;574(7777):273-277.
- Trivedi G, Inoue D, Chen C, Bitner L, Chung YR, Justin T, Gönen M, Wess J, Abdel-Wahab O, and Zhang L. Muscarinic acetylcholine receptor regulates self-renewal of early erythroid progenitors. Sci Transl Med. 2019 Sep 25;11(511).
- 12. Taylor J, Sendino M, Gorelick AN, Pastore A, Chang MT, Penson AV, Gavrila EI, Stewart C, Melnik EM, Herrejon Chavez F, Bitner L, Yoshimi A, Lee SC, Inoue D, Liu B, Zhang XJ, Mato AR, Dogan A, Kharas MG, Chen Y, Wang D, Soni RK, Hendrickson RC, Prieto G, Rodriguez JA, Taylor BS, Abdel-Wahab O. Altered nuclear export signal recognition as a driver of oncogenesis. Cancer Discov, 2019 Oct;9(10):1452-1467.
- Wang E, Lu SX, Pastore A, Chen X, Imig J, Chun-Wei Lee S, Hockemeyer K, Ghebrechristos YE, Yoshimi A, Inoue D, Ki M, Cho H, Bitner L, Kloetgen A, Lin KT, Uehara T, Owa T, Tibes R, Krainer AR, Abdel-Wahab O, Aifantis I. Targeting an RNA-Binding Protein Network in Acute Myeloid Leukemia. Cancer Cell. 2019 Mar 18;35(3):369-384.
- Saika M, Inoue D, Nagase R, Sato N, Tsuchiya A, Yabushita T, Kitamura T, Goyama S. ASXL1 and SETBP1 mutations promote leukaemogenesis by repressing TGFβ pathway genes through histone deacetylation. Sci Rep. 2018 Oct 26;8(1):15873.
- Lee SC, North K, Kim E, Jang E, Obeng E, Lu SX, Liu B, Inoue D, Yoshimi A, Ki M, Yeo M, Zhang XJ, Kim MK, Cho H, Chung YR, Taylor J, Durham BH, Kim YJ, Pastore A, Monette S, Palacino J, Seiler M, Buonamici S, Smith PG, Ebert BL, Bradley RK, Abdel-Wahab O. Synthetic Lethal and Convergent Biological Effects of Cancer-Associated Spliceosomal Gene Mutations. Cancer Cell. 2018 Aug 34(2):225-241.
- Asada S, Goyama S, InoueD, Shikata S, Takeda R, Fukushima T, Yonezawa T, Fujino T, Hayashi Y, Kawabata KC, Fukuyama T, Tanaka Y, Yokoyama A, Yamazaki S, Kozuka-Hata H, Oyama M, Kojima S, Kawazu M, Mano H, Kitamura T. Mutant ASXL1 cooperates with BAP1 to promote myeloid leukaemogenesis. Nat Commun. 2018 Jul16;9(1):2733.
- Taylor J, Pavlick D, Yoshimi A, Marcelus C, Chung SS, Hechtman JF, Benayed R, Cocco E, Durham BH, Bitner L, Inoue D, Chung YR, Mullaney K, Watts JM, Diamond EL, Albacker LA, Mughal TI, Ebata K, Tuch BB, Ku N, Scaltriti M, Roshal M, Arcila M, Ali S, Hyman DM, Park JH, Abdel-Wahab O. Oncogenic TRK fusions are amenable to inhibition in hematologic malignancies. J Clin Invest. 2018 Aug 31;128(9):3819-3825.
- Nagase R, Inoue D (co-first and Corresponding author), Pastore A, Fujino T, Hou HA, Yamasaki N, Goyama S, Saika M, Kanai A, Sera Y, Horikawa S, Ota Y, Asada S, Hayashi Y, Kawabata KC, Takeda R, Tien HF, Honda H, Abdel-Wahab O, Kitamura T. Expression of mutant Asxl1 perturbs hematopoiesis and promotes susceptibility to leukemic transformation. J Exp Med. 2018 Jun 215(6):1729-1747.
- Inoue D (Corresponding author), Fujino T, Sheridan P, Zhang YZ, Nagase R, Horikawa S, Li Z, Matsui H, Kanai A, Saika M, Yamaguchi R, Kozuka-Hata H, Kawabata KC, Yokoyama A, Goyama S, Inaba T, Imoto S, Miyano S, Xu M, Yang FC, Oyama M, Kitamura T. A novel ASXL1-OGT axis plays roles in H3K4 methylation and tumor suppression in myeloid malignancies. Leukemia. 2018 Jun 32(6):1327-1337.
- Kawabata KC, Hayashi Y, Inoue D, Meguro H, Sakurai H, Fukuyama T, Tanaka Y, Asada S, Fukushima T, Nagase R, Takeda R, Harada Y, Kitaura J, Goyama S, Harada H, Aburatani H, Kitamura T. High expression of ABCG2 induced by EZH2 disruption plays pivotal roles in MDS pathogenesis. Leukemia. 2018 Feb 32(2):419-428.
- Inoue D, Fujino T, Kitamura T. ASXL1 as a critical regulator of epigenetic marks and therapeutic potential of mutated cells. LOncotarget. 2018 Oct 16;9(81):35203-35204.
- Micol JB, Pastore A, Inoue D (co-first author), Duployez N, Kim E, Lee SC, Durham BH, Chung YR, Cho H, Zhang XJ, Yoshimi A, Krivtsov A, Koche R, Solary E, Sinha A, Preudhomme C, Abdel-Wahab O. ASXL2 is essential for haematopoiesis and acts as a haploinsufficient tumour suppressor in leukemia. Nat Commun. 2017 May 18;8:15429, 2017.
- Inoue D, Abdel-Wahab O. Modeling SF3B1 Mutations in Cancer: Advances, Challenges, and Opportunities. Cancer Cell. 2016 Sep 12;30(3):371-373.
- Inoue D, Bradley RK, Abdel-Wahab O. Spliceosomal gene mutations in myelodysplasia: molecular links to clonal abnormalities of hematopoiesis. Genes Dev. 2016 May 1;30(9):989-1001.
Major Awards in Recent Years
(Daichi Inoue)
- • UJA outstanding publication award, United Japanese Researchers Around the World
- • Inoue Research Award, Inoue Foundation for Science
- • Institute of Biomedical Research and Innovation Director's Award, Foundation for Biomedical Research and Innovation at Kobe
- • Foundation for Biomedical Research and Innovation at Kobe President's Award, Foundation for Biomedical Research and Innovation at Kobe
- • ASH (American Society of Hematology) Global Research Award, American Society of Hematology
- • Kirin-ji Award, Kirin Juku 2021




